Research Article- Asian Journal of Pharmaceutical Technology and Innovation(2018)
Proteomic analysis to determine mechanism of purple rice bran on pancreatic damage of type 2 diabetic rats
Ei Ei Hlaing1, Supicha Rungjarernarreejitt2, Puriwat Fakfum1, Patipat Kamdenlek1 and Pichapat Piamrojanaphat1*2Department of Biochemistry, University of Medicine, Mandaley, Myanmar
Dr. Pichapat Piamrojanaphat, Department of Biochemistry, Faculty of Medicine, Chiang Mai University, Thailand, Tel: +66 53 941000, Email: phichapat.p@cmu.ac.th
Received: 25-May-2018 Accepted Date: Jun 01, 2018 ; Published: 07-Jun-2018
Abstract
Diabetes is one of the most common metabolic diseases all over the world. Type 2 diabetic contributes nearly 90% of diabetic patients. Type 2 diabetes is characterized by hyperglycemia with dysfunction of pancreatic beta cells and insulin resistance. Anthocyanins are the major functional component in purple rice (Oryza sativa L. indica). Our previous studies showed that supplementation of purple rice husk attenuated hyperglycemia and prevented pancreatic damage. However, the molecular mechanism of anti-diabetes activities of purple rice need more investigated. In this study, pancreatic tissue samples of type 2 diabetic rats with or without purple rice bran supplement were studied through proteomic approach. Five unique proteins were found in purple rice bran supplemented diabetic rats (Rasgrp3, Clca 1, Olr3, Abcc4, Mfn1). These 5 proteins interacted with other proteins functioning in glycolysis (Eno 3) and insulin signaling (Irs 1, Irs 2, Foxo 1, Foxo 3). Through western blot analysis, Glut 2 was restored back in purple rice bran supplementation, indicating the purple rice bran increase glucose uptake of pancreas and increase the release of insulin. Interestingly, glucose transporter 4 (Glut 4) protein were restored back in purple rice bran supplemented diabetic rats via the expression of Myosin I heavy chain (Myo 1c). Moreover, unique protein Rasgrp 3 might promote pancreatic β-cell proliferation via Ras/MEK pathway together with phosphorylation of Akt protein. This is the first study to show the molecular mechanism of purple rice bran on pancreatic tissue of diabetes rats. All of the results suggested that purple rice bran could increase glucose uptake, insulin signaling and β-cells survival.
Introduction
Diabetes is one of the most common metabolic diseases all over the world. International Diabetes Federation (IDF) estimated that prevalence of diabetes continues to rise reaching 642 million by 2040 (1). In type 2 diabetes, the body builds up resistance to insulin and more insulin is needed to bring down blood glucose levels. As a result the pancreas needs to produce more insulin than it would normally need to. If the pancreas can no longer produce enough insulin to bring down sugar levels, the symptoms of diabetes will begin to appear. Type 2 diabetes comes on gradually and it can take up to years for symptoms to appear. Further development of type 2 diabetes can lead to loss of insulin producing beta cells from the pancreas (2).
The principal features of type 2 diabetes are an abnormal sensitivity of peripheral tissues to insulin and decreased levels of hormone secretion. In pancreatic β cells, glucose oxidation and lipid peroxidation always lead to reactive oxygen species production, and which are normally detoxified by catalase and superoxide dismutase. However, β cells are composed of small amount of these proteins and glutathione peroxidase (antioxidant enzyme). Excess of glucose and lipid lead to formation of large amount of reactive oxygen species in β cells, followed by damage to cellular components. Additionally, reactive oxygen species are known to enhance NF-KB activity, which potentially induces β cell apoptosis (3).
In the presence of glucose, fatty acid oxidation in β cells is inhibited. As a result, there will be accumulation of long chain acyl coenzyme A, which has been proved to be a part of normal insulin secretion process. However, on the other hand, long-chain acyl coenzyme A can also diminish the insulin secretion by opening potassium channels of β cells. Another possible mechanism is the reduction insulin secretion by inhibiting Akt activity. Akt overexpressed mice increased insulin secretion together with higher beta cell mass (4). Conversely, a kinasedead mutant of Akt transgenic mice decrease in glucose stimulated insulin secretion (5). Another proposed mechanism is that an increased expression of uncoupling protein 2 which would lead to decrease ATP production from respiratory chain and reduction in insulin secretion. Reduction in insulin secretion, oxidative stress, mitochondrial dysfunction and apoptosis leads to β cells dysfunction and reduction in cell mass.
Purple rice (Oryza sativa L. indica) is cultivated in Southeast Asia, including Thailand, China and Indonesia. Major functional component of pigmented rice is anthocyanins. Anthocyanins have been proved to have antioxidant, anti-inflammatory, anti-diabetes and anti-hypercholesterolemia. Our previous study also showed that anthocyanin-rich bran from Thai purple rice genotype Kum Doi Saket ameliorated hyperglycemia and hyperlipidemia in high-fat diet plus STZ induced type 2 diabetic rats. We revealed that supplementation of purple rice extract not only attenuated hyperglycemia, but also prevented the rise in pancreatic lipid peroxidation and enhanced the pancreatic antioxidant defense capacity (6). However, the molecular mechanism has not been investigated. Nowadays, proteomics study becomes the interesting research scope for analyzing different protein expressions in different diseases. It is used to explore molecular pathways and find biomarkers. Therefore, the present study, the proteomic technique was performed to investigate whether the antidiabetic effect of anthocyanins-rich bran from Thai purple rice was associated with the improvement of pancreatic structure and function in type 2 diabetic.
Materials and Methods
Tissue Preparation and Homogenization
In this study, twenty adults male wistar rats were provided by Dr. Narissara Lailerd and colleagues from Department of Physiology, Faculty of Medicine, Chiang Mai University (protocol number: 27/2558). Pancreatic tissues were sacrificed from four group of five rats each including Group I: normal rats control (NDC); Group II: normal rats receiving normal diet supplemented with purple rice bran at a dose of 50 g /kg diet (NDR); Group III: type 2 diabetic rats control (DM2C); Group IV: type 2 DM rats received high-fat diet plus purple rice bran 50 g /kg diet (DM2R).
Pancreas tissues were pulverized in liquid nitrogen using mortar and pestle. Then fine powder was dissolved in 1 ml of 0.5% SDS. The mixture was centrifuged at 10,000 g for 15 minutes. The supernatant was collected and mixed with 2 volumes of cold acetone, and then placed at -20˚C overnight. The following day, the mixture was centrifuged at 10,000 g, 15 min and got the protein pellet. Finally, the protein pellet was resuspended with 0.5% SDS and stored at -20˚C. Protein concentration in each tissue samples was determined by Lowry assay using bovine serum albumin (BSA) as standard. The proteins from each sample was adjusted to an equal protein concentration and pooled together according to groups for further SDS-PAGE analysis.
One Dimensional SDS-PAGE Analysis (1D SDS PAGE)
In SDS-PAGE, proteins were separated based on polypeptide length, and so their molecular weight. After proteins are separated by SDS-PAGE, then the gels were stained with silver staining.
The in-gel digestion is part of the sample preparation for the mass spectrometric identification of proteins. The excised gel pieces was in gel digested using an in-house protocol developed in National Center for Genetic Engineering and Biotechnology (BIOTEC), Bangkok, Thailand (7).
Peptide Identification and Quantitation by LC-MS/MS Analysis
For protein quantitation, DeCyder MS 2.0 Differential Analysis software (GE Healthcare, Sweden) (8,9) was used. Acquired MS/MS raw data were converted to mzXML files using CompassXport 1.3.10 (Bruker, Germany) prior to import into DeCyder MS software.
The analyzed MS/MS data from DeCyder MS were submitted to database search using the Mascot software version 2.2 (Matrix Science, UK) (10). The data were searched against the NCBI database for protein identification.
Bioinformatics Analysis
The information (such as molecular weight, molecular function, structure, sequence) of interested proteins from Mascot search was searched against Uniprot database (http://www.uniprot.org/). Search tool for the retrieval of interacting genes/proteins (STRING version 10) were used to propose possible mechanism(s) of diabetes and anti-diabetes effects of purple rice bran.
Validation of Proteins Expression level by Western Blot Analysis
To propose the molecular mechanism(s) of antidiabetic activities of purple rice bran, phosphorylated p-Akt Ser473, Glut 2, and Glut 4 protein expression in pancreatic tissue were measured. Image J software was used to detect the band density. Protein concentration was compared with the normal group, setting as arbitrarily 100.
Statistical Analysis
The experimental data were shown as mean ± SE or mean ± SD as appropriate. Statistical analysis was done by analysis of variance (ANOVA) and followed by post hoc LSD test to determine specific mean difference. A significant difference was set at p <0.05.
Results
Identification of Pancreatic Proteins
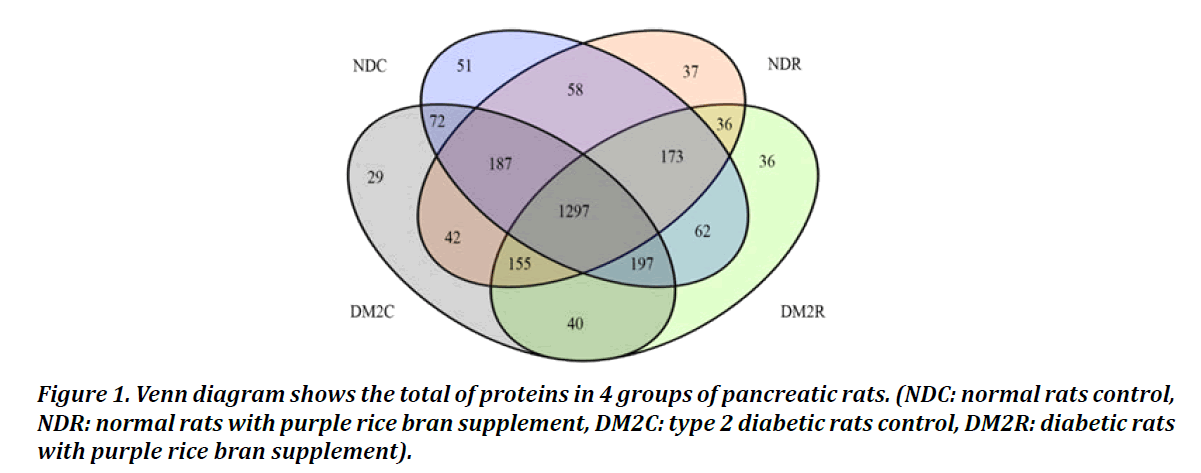
2472 proteins were differentially expressed proteins in all four group as shown in Figure 1. The numbers of the identified proteins in NDC, NDR, DM2C, and DM2R were 2097,1985,2019 and 1996 respectively. Total of 1297 proteins were found in all four groups consist of 51, 29, and 36 unique proteins in the NDC, DM2C, and DM2R group respectively.
Bioinformatic Analysis of Unique Proteins Found in The Pancreatic Tissues of Diabetic rats and Diabetic rats
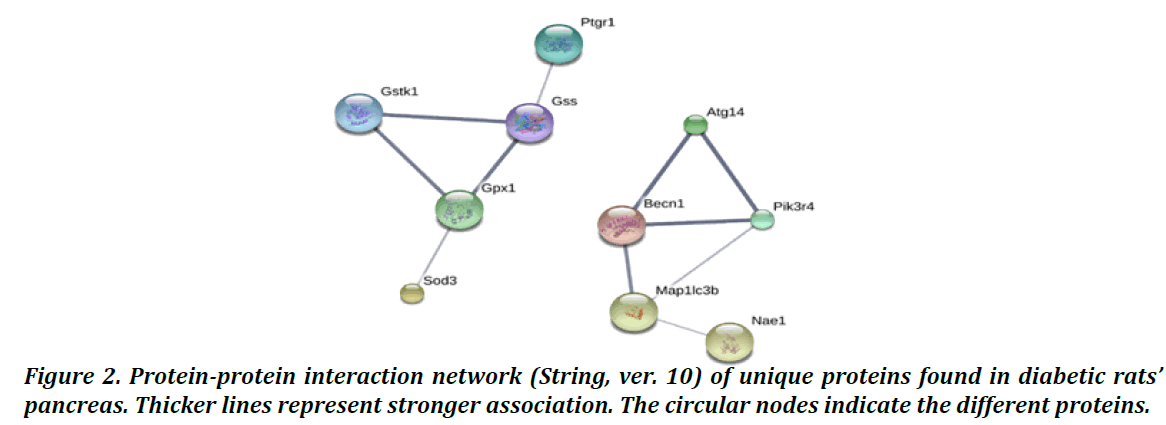
To get more information regarding the molecular mechanism of diabetes, we constructed the protein interaction network between the unique proteins of the DM2C group and other proteins by using the STRING (ver.10) software. The result was as shown in Figure 2, six additional interacting proteins (Becn1, Pik3r4, Gss, Gpx1, Gstk1 and SOD3) were put in the network construction to get more perfect pathway. Interestingly, 4 out of 14 unique proteins (Nae1, Atg14, Ptgr1 and Map1l3cb) (Table 1) were found to be associated with proteins related with oxidative stress response proteins (Gss, Gpx1, Gstk1 and SOD3) and proteins related with autophagy pathway (Becn1, and Pik3r4). This will support the involvement of pancreatic β cell death in the development of diabetes.
| Accession No. | Protein name | Gene name | Functional categories | Peptide sequence |
|---|---|---|---|---|
| gi50400224 | NEDD8-activating enzyme E1 regulatory subunit | Nae1 | Autophagy | EKEDFR |
| gi32264635 | Zbs559 | Map1lc3b | Autophagy | TLHSLY |
| gi157822327 | Beclin 1 associated autophagy related key regulator | Atg14 | Autophagy | EVLKAMEGK |
| gi20302022 | Prostaglandin reductase 1 | Ptgr1 | oxidoreductase | VVESK |
Table 1: Identification and functional categories of 4 unique proteins found in diabetic rats’ pancreas.
Bioinformatic Analysis of Unique Proteins Found in The Pancreatic Tissues of Diabetic rats With Purple Rice Bran Supplement
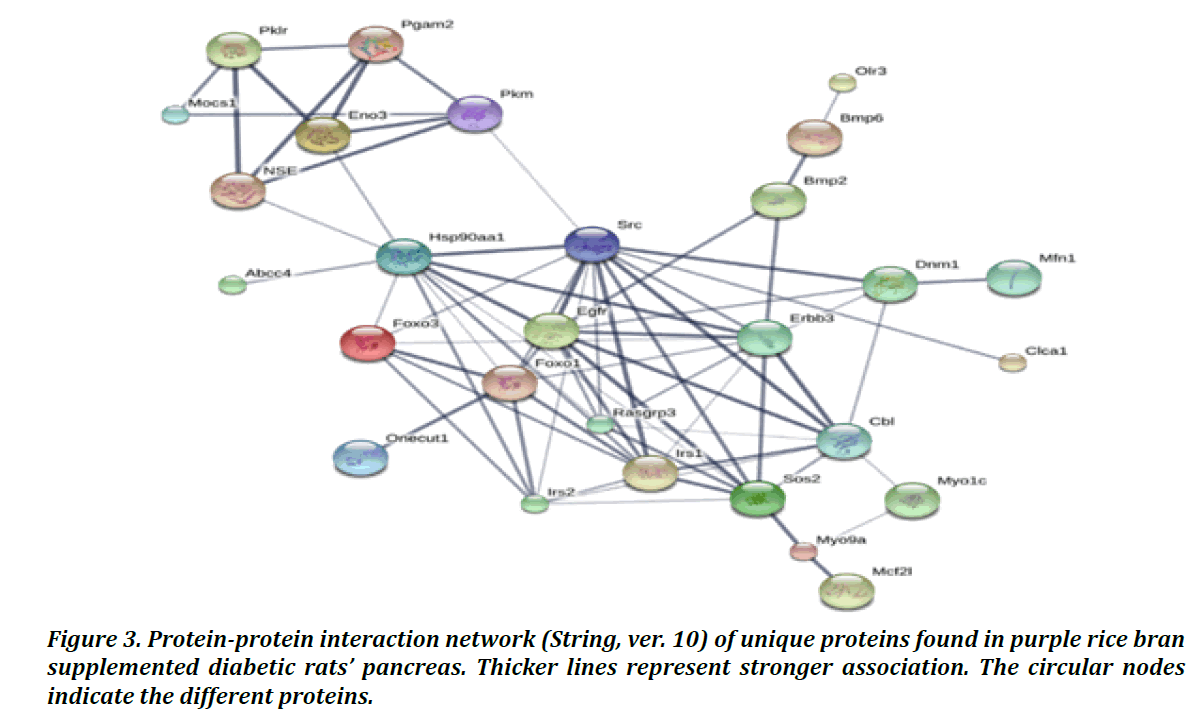
The protein interaction network of unique protein in DM2R rats’ pancreas was studied by using the STRING (ver.10) database. As shown in Figure 3, five unique proteins (Rasgrp3, Clca 1, Olr3, Abcc4, Mfn1) (Table 2) were found to be connected with other proteins functioning in glycolysis (Pklm, Pgam2, Pkm, Eno3) and insulin signaling (such as Irs1, Foxo1, Foxo3, Irs 2). Interestingly, myosin I heavy chain (Myo 1c) (gi400429) is a protein that function in Glut 4 translocation and it was upregulated in both NDR and DM2R rats’ pancreatic protein of this study. Furthermore, Myo 1 c showed a correlation with unique DM2R protein in STRING network. Based on this information, purple rice bran supplement might involve in insulin signaling and glucose metabolism.
| Accession No. | Protein name | Gene name | Functional categories | Peptide sequence |
|---|---|---|---|---|
| gi157818579 | Calcium-activated chloride channel regulator1 precursor | Clca1 | Cell membrane | QGASPILR |
| gi157819575 | Ras guanyl-releasing protein 3 | Rasgrp3 | Cell signaling | DKGFAK |
| gi20376820 | Mitofusin-1 | Mfn1 | cytoskeleton | NPELDR |
| gi4757727 | Olfactory receptor | Olr3 | Cell signaling | AAFGR |
| gi1416570 | Multidrug resistance protein 4 | Abcc4 | Transporter | NRILIIDEATANVDPR |
| gi58865398 | Cytosol aminopeptidase | Lap3 | Protein catabolism | QVIDCQLADVNNLGK |
Table 2: Identification and functional categories of 23 unique proteins found in purple rice bran supplemented diabetic rats’ pancreas.
Validation of Proteins Expression Level from Pancreatic Proteomic Analysis
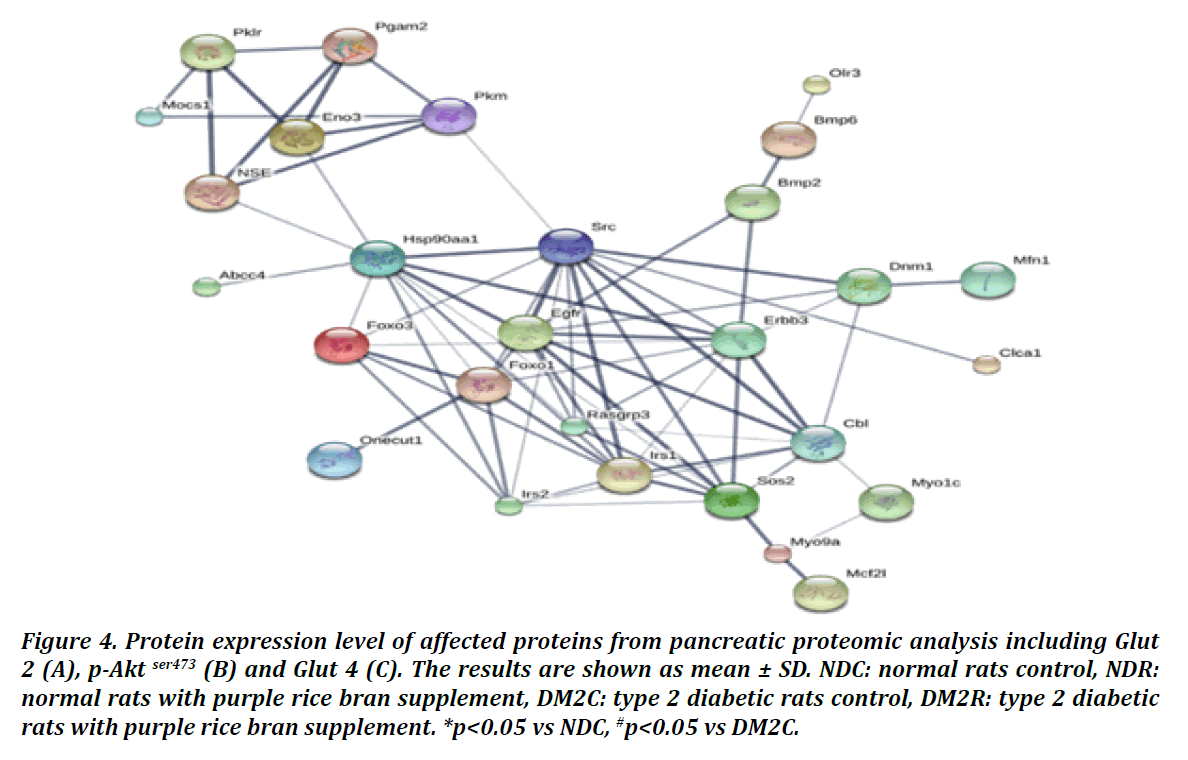
To determine the expression of proteins that functioning in insulin signaling and glucose uptake. Protein expression level of Glut 2, p-Akt ser 473, and Glut 4 were determined by western blot analysis. In this study found that Glut 2 expression was significantly reduced in DM2C group when comparison with NDC. Moreover, it was significantly increased in DM2R when compare with DM2C. However, Glut 2 expression in DM2R was comparable with NDC rats (Figure 4A). p- Akt ser 473 expression was significantly increased DM2R when compared with DM2C but no different between the experimental groups (Figure 4B). Glut 4 expression was significantly decreased in DM2C rats when compared with NDC and DM2R showing in Figure 4. From these data indicated that purple rice bran can improve glucose uptake and insulin signaling in diabetic rats.
Figure 4: Protein expression level of affected proteins from pancreatic proteomic analysis including Glut 2 (A), p-Akt ser473 (B) and Glut 4 (C). The results are shown as mean ± SD. NDC: normal rats control, NDR: normal rats with purple rice bran supplement, DM2C: type 2 diabetic rats control, DM2R: type 2 diabetic rats with purple rice bran supplement. *p<0.05 vs NDC, #p<0.05 vs DM2C.
Discussion
Type 2 diabetes is characterized by insulin resistance in peripheral tissue and impaired insulin secretion from pancreatic-β-cells dysfunction and/or the reduction of β cells mass. (11).
Pancreatic β cells loss in type 2 diabetes results from apoptosis, oxidative stress and endoplasmic reticulum stress. Based on those studies, β cell loss has been reported to be a participant in the onset of diabetes. In this study, guanyl nucleotide-releasing protein for Ras 3 (Rasgrp 3) found in type 2 diabetic rats supplement with purple rice bran. Rasgrp 3 is a member of the RasGRP family that activated MAP kinase kinase kinase phosphorylation (Raf) (12). It has previously reported that Raf 1 is required for β-cell survival and proliferation (13). From this finding, it can be summarized that purple rice bran can affect to Rasgrp3 expression to increase pancreatic β-cell proliferation and β-cell survival. According STITCH 4.0, we found interaction of Rasgrp3 with Myo 1c that associated with Glut 4 translocation.
Myo 1 c, myosin I heavy chain, is an actin-based motor protein. It facilitates insulin-stimulated Glut 4 translocation in adipocyte and skeletal muscle. Previously, in 3T3-L1 adipocytes, the high expression Myo1c promoted the insulin stimulated Glut 4 transport in adipocytes (14). Moreover, there was enhanced glucose uptake in skeletal muscles expressing wild-type Myo1c and impaired glucose uptake in skeletal muscles expressing mutant Myo1c. And they proposed that Myo1c increase glucose uptake by transporting Glut 4 to cellular membrane (15). However, Glut 4 is not a major glucose transporter in pancreas and Myo1 c expression in pancreas had not been reported. Interestingly, we found that Myo1c expressed in pancreatic tissue of purple rice bran supplemented diabetic group.
Glucose transporter 2 (Glut 2) is the principal glucose transporter in pancreatic tissue. Even though, glucose transporter 4 (Glut 4) also expressed in pancreatic tissue. Phosphorylated Akt (p-Akt), from insulin signaling, promotes Glut 4 translocation to membrane. From western blot analysis, purple rice bran restored the Glut 4 expression which was reduced in diabetic rats. In addition, Glut 2 protein expression is significantly increased in purple rice bran supplement in type 2 diabetic rats supplement with purple rice bran when compared with diabetic rats, it can be summarized that purple rice bran activated Glut 2 and Glut 4 expression that stimulate glucose uptake in pancreas and reduce blood glucose level (16). Moreover, purple rice bran enhances glucose sensing system in diabetic rats and increased glucose uptake via Myo 1c promoting Glut 4 trafficking.
PI3K/Akt pathway is cell signaling pathway that regulate pancreatic β cell mass and proliferation. In previously studies found that activation of Akt signaling in transgenic mice increased β-cell mass and proliferation and maintained glucose- stimulated insulin secretion (17). Meanwhile, reduction of Akt activity in transgenic animals resulted in impaired glucose tolerance due to defective insulin secretion disturbing at the insulin exocytosis mechanism (18). From western blot analysis, purple rice bran increased phosphorylated Akt expression in pancreas. It can be suggested that Akt regulate pancreatic cell growth and maintain insulin secretion.
Conclusion
To date, there have been several studies on local purple rice focusing on anti-oxidant, anti-inflammation and anti-diabetic activity. However, there is not any study reveal the molecular mechanism of purple rice or purple rice bran on pancreatic tissue of diabetic rats. Because this study showed that purple rice bran can regulate β cell mass, β cell-survival, improve insulin signaling and glucose homeostasis. This purple rice bran is considered to be a great source of anti-diabetes, it may be useful in prevention and treatment of pancreatic damage in human with risk of diabetes and diabetes patients. However, the mechanism of purple rice bran still need more investigate. Significantly more research is required in working focus on the nutraceutical value and toward the development of purple rice bran as a drug or functioning food.
Acknowledgements
This study supported by Faculty of Medicine, Chiang Mai University. We wish to thank Dr. Sittiruk Roytrakul at National Center for Genetic Engineering and Biotechnology (BIOTEC) and National Research University Project under Thailand’s office of the Higher Education Commission.
Conflict of Interest
The authors have no conflict of interest.
References
- Jaacks LM, Siegel KR, Gujral UP, Narayan KMV, Type 2 diabetes: A 21st century epidemic, Best Practice & Research Clinical Endocrinology & Metabolism, 30 (3); 331-343, 2016.
- Diagnosis and Classification of Diabetes Mellitus, Diabetes Care.014; 37(Supplement 1):S81.
- Robertson RP, Harmon J, Tran PO, Tanaka Y, Takahashi H, Glucose Toxicity in β Cells: Type 2 Diabetes, Good Radicals Gone Bad, and the Glutathione Connection, Diabetes, 52 (3); 581, 2003.
- Bernal-Mizrachi E, Wen W, Stahlhut S, Welling CM, Permutt MA, Islet β cell xpression of constitutively active Akt1/PKBα induces striking hypertrophy, hyperplasia, and hyperinsulinemia, Journal of Clinical Investigation, 108 (11); 1631-1638, 2001.
- Bernal-Mizrachi E, Fatrai S, Johnson JD, Ohsugi M, Otani K, Han Z, Polonsky KS, Permutt MA, Defective insulin secretion and increased susceptibility to experimental diabetes are induced by reduced Akt activity in pancreatic islet β cells. Journal of Clinical Investigation, 114 (7); 928-936, 2004.
- Intakhad JTP, Kobroob A, Keapai W, Wongpoomchai R, Wongmekiat O, Lailerd N, Therapeutic Efficacy of Purple Rice Husk Extraction Pancreatic Damage in High Fat-Streptozotocin Induced Diabetic Rats Proceeding of the 19th national graduate research conference, 899-907, 2018.
- Abere B, Wikan N, Ubol S, Auewarakul P, Paemanee A, Kittisenachai S, Sittiruk Roytrakul ,Duncan R, Proteomic analysis of chikungunya virus infected microgial cells, PLoS One, 7(4); e34800, 2012.
- Johansson C, Samskog J, Sundstrom L, Wadensten H, Bjorkesten L, Flensburg J, Differential expression analysis of Escherichia coli proteins using a novel software for relative quantitation of LC-MS/MS data, Proteomics, 6 (16); 4475-85, 2006.
- Thorsell A, Portelius E, Blennow K, Westman‐Brinkmalm A, Evaluation of sample fractionation using micro‐scale liquid‐phase isoelectric focusing on mass spectrometric identification and quantitation of proteins in a SILAC experiment. Rapid Communications in Mass Spectrometry, 21 (5); 771-778, 2007.
- Cottrell JS, London U, Probability-based protein identification by searching sequence databases using mass spectrometry data, Electrophoresis, 20 (18); 3551-3567, 1999.
- Butler AE, Janson J, Bonner-Weir S, Ritzel R, Rizza RA, Butler PC, Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes, Diabetes, 52 (1); 102-110, 2003.
- Ward AF, Braun BS, Shannon KM, Targeting oncogenic Ras signaling in hematologic malignancies. Blood, 120 (17); 3397-3406, 2012.
- Beith JL, Alejandro EU, Johnson JD, Insulin stimulates primary beta-cell proliferation via Raf-1 kinase, Endocrinology, 149 (5); 2251-2260, 2008.
- Bose A, Guilherme A, Robida SI, Nicoloro SM, Zhou QL, Jiang ZY, Pomerleau DP, Czech MP, Glucose transporter recycling in response to insulin is facilitated by myosin Myo1c. Nature, 420 (6917); 821-824, 2002.
- Toyoda T, An D, Witczak CA, Koh HJ, Hirshman MF, Fujii N, Goodyear LJ, Myo1c regulates glucose uptake in mouse skeletal muscle, J Biol Chem, 286 (6); 4133-4140, 2011.
- Arya A, Al-Obaidi MM, Karim RB, Taha H, Khan AK, Shahid N, Sayem AS, Looi CY, Mustafa MR, Mohd MA, Ali HM, Extract of Woodfordia fruticosa flowers ameliorates hyperglycemia, oxidative stress and improves beta-cell function in streptozotocin-nicotinamide induced diabetic rats, J Ethnopharmacol, 175; 229-240, 2015.
- Bernal-Mizrachi E, Wen W, Stahlhut S, Welling CM, Permutt MA, Islet beta cell expression of constitutively active Akt1/PKB alpha induces striking hypertrophy, hyperplasia and hyperinsulinemia, J Clin Invest, 108 (11); 1631-1638, 2001.
- Bernal-Mizrachi E, Fatrai S, Johnson JD, Ohsugi M, Otani K, Han Z, Polonsky KS, Permutt MA, Defective insulin secretion and increased susceptibility to experimental diabetes are induced by reduced Akt activity in pancreatic islet beta cells, J Clin Invest, 114 (7); 928-936, 2004.
Citation: Ei Ei Hlaing, Supicha Rungjarernarreejitt, Puriwat Fakfum, Patipat Kamdenlek, Pichapat Piamrojanaphat, Proteomic analysis to determine mechanism of purple rice bran on pancreatic damage of type 2 diabetic rats, 6(28); 22-38, 2018.. www.asianpharmtech.com.